Computational method development for human immunogenetics
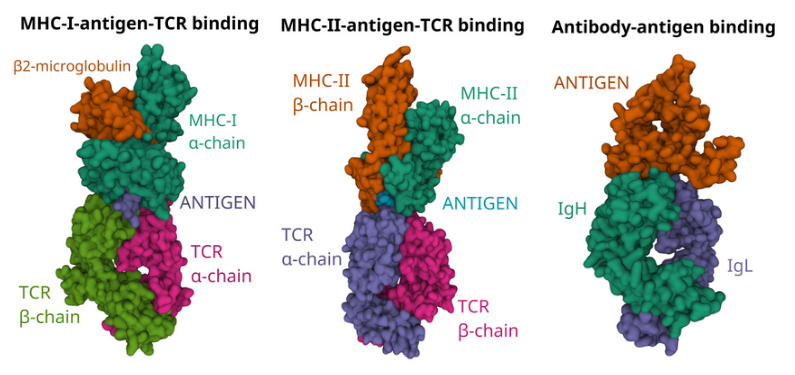
Immune molecules such as B and T cell receptors, human leukocyte antigens (HLAs) and killer Ig-like receptors (KIRs) are encoded in the most diverse genetic loci in the human genome. Bioinformatic methods commonly used to analyse immune responses in large patient cohorts do not take into account this immunogenetic diversity, which leads to erroneous quantification of important immune mediators and impaired inter-donor comparability. Our research group develops robust bioinformatic methods and software tools for reproducible research on human immune responses and immunogenetic diversity in multidimensional immunological datasets.
We greatly acknowledge funding from the Single Cell Program of the Chan Zuckerberg Foundation.
Packages
immunotation — R/Bioconductor package
This package provides tools for consistent annotation of HLA genes in typical immunoinformatics workflows such as for example the prediction of MHC-presented peptides in different human donors.
SingleCellAlleleExperiment — R package
This package is a container for storing and handling allele-aware quantification data for immune genes at a multi-layer level.


